Segmental Duplication NEAT (SDNEAT)
Segmental Duplication NEAT is based on NEAT and inspired by recent research of the human genome. This recent research claims to show that large segments of the human genome that are purely duplicate genetic information may be critical requirements for the advancement of the species.
Nearly 14% of the human genome consists of segmental duplications. In comparison, the mouse genome is approximately 7% segmental duplications and the chimpanzee genome is only about 5% segmental duplications. These segmental duplications also appear to be at least somewhat non-random. Segmental duplications show a higher rate of copy variation, or mutation. They appear to be favoured in gene selection and several functional categories that are realized in human beings appear to be enriched by segmental duplications. One example of this enrichment is the human immune system. The high percentage of segmental duplications in the human genome seems to imply that they are key to faster innovation through genetic processes. They protect the genome from harmful mutations because they are duplicates, and most mutations to them will not affect the existing genomic functionality. The human male gender chromosome (the ‘Y’ chromosome), shows a very high amount of segmental duplications; approximately 50% of its genes are segmental duplications. This may imply that segmental duplications prevent genetic stagnation in the male of the species; the ‘Y’ chromosome routinely undergoes mutation. This amount of mutation is required as the ‘Y’ chromosome never performs crossover with other chromosomes.
All these reasons support the development of a new version of the NEAT algorithm. Segmental Duplication NEAT (SDNEAT) is be based on the NEAT algorithm but includes a new mutation operator which will identify a segment of genetic information, duplicate that segment, heavily mutate it, and integrate it back into the genome. This duplicated segment may offer an evolutionary leap, and cause the algorithm to find new solutions to the problem. Using SDNEAT, innovations are still protected by speciation so all the advantages of the NEAT algorithm are preserved. It is important to note that NEAT would be capable of evolving any solution SDNEAT can evolve. However the chances of NEAT evolving exactly the same segmental duplication are quite low as it would require multiple new node innovations in a particular sequence.
A Segment
In order to develop a segmental duplication operation a segment must first be defined, because the concept of segments does not exist in the original NEAT algorithm.
This definition states that all segments for SDNEAT begin at an input node, end at an output node and must contain at least one hidden node to a maximum of n hidden nodes. There are no recurrent or loopback connections in a valid segment. Some of the problems of identifying a valid segment algorithmically are eliminated by limiting a valid segment to this subset. The portion of the algorithm that identifies a valid segment need only walk a path through the neural network from an input node to an output node. It can ignore recurrent connections along the way.
Segmental Duplication
Because identification of a segment is simple and the beginning and endpoint of a segment is limited to an input node and an output node, inserting the duplicated segment is also straightforward. The identified segment is already a valid path in the neural network so duplicating it and inserting it between the same input and output nodes does not destroy the genome, but it does modify a substantial portion of the genome’s genetic code. This enhanced rate of growth does not significantly increase complexity as it relies on the original NEAT methods for topological growth and cannot evolve any structure that NEAT could not. It simply causes generational leaps to happen faster. In fact, no segmental duplications can occur without original NEAT node mutations. The initial genomes contain only input and output nodes and because, by definition, a segment cannot contain an input and output node.
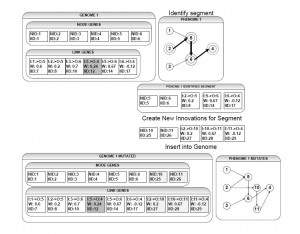
Figure 1: Segmental Duplication. In Genome One at the top of this diagram, a segment has been identified. The nodes with solid circles are added to S_n and the highlighted links connecting the nodes with broken circles surrounding them are added to L_m. This segment is then duplicated and new innovation numbers are created for all the components, the node identifiers are properly incremented and the links are adjusted to connect to the new nodes. These nodes and links are then appended to Genome One and the new phenome is displayed at bottom right.
The historical markings or innovation numbers are an important aspect of NEAT. When SDNEAT inserts a new segmental duplication, it is creating a copy of active genes. In SDNEAT, all segmental duplications are treated as new innovations. In the original NEAT algorithm, if a node mutation occurs which disables a link, then later that link is re-enabled and an equivalent node mutation occurs on the same link the innovation list identifies this as an old innovation. Although the innovation list has identified it as an old innovation, NEAT considers it to be a new innovation and has the innovation list assign it a new innovation number. This is the base case of a segmental duplication, and consequently all segmental duplications are treated as new innovations.
To identify a segment, the SDNEAT algorithm first randomly selects an input node from the genomes set of input nodes. Then the algorithm attempts to find a path to an output node, randomly chooses an output link from its current node and, if the output link is not recurrent, the algorithm follows that link to the next node and repeats the previous steps. Each time the algorithm steps to a new node it copies the link and node to its arrays for duplication. If a step from an input node arrives at an output node of the neural network, a new input node is randomly chosen and the algorithm starts again, as by definition a segmental duplication cannot consist of one link. If the algorithm finds an output node, it has found a valid segment. The algorithm duplicates the valid segment by creating new innovations for each link and node in the segment’s link and node arrays. The weights from the original nodes and links are duplicated but the innovation numbers are updated. The segment’s weights are then mutated at a higher than average mutation rate. Once mutation of the segment is complete the new links and nodes are appended to the genome being mutated.
SDNEAT maintains the efficiencies and capabilities of NEAT, including all operations and speciation, but it introduces a new operator: the segmental duplication mutation. This new operator can drastically mutate an existing genome without affecting the capabilities of the existing NEAT algorithm. This drastic mutation has the potential to broaden the search area of NEAT to include elements that would not otherwise be searched for several generations. This mimics the genetic behaviour recently identified in the human genome. In order to evaluate this new algorithm and its ability to evolve an efficient neural network controller that can learn to effectively operate an autonomous agent in multiple different dynamic environments, a neuroevolutionary simulation system must be built.